
Drive innovation with our technology github - bob-friedman protein-sequence-generation: python code to help gallery of extensive collections of digital images. technologically showcasing photography, images, and pictures. ideal for innovation showcases and presentations. Our github - bob-friedman protein-sequence-generation: python code to help collection features high-quality images with excellent detail and clarity. Suitable for various applications including web design, social media, personal projects, and digital content creation All github - bob-friedman protein-sequence-generation: python code to help images are available in high resolution with professional-grade quality, optimized for both digital and print applications, and include comprehensive metadata for easy organization and usage. Our github - bob-friedman protein-sequence-generation: python code to help gallery offers diverse visual resources to bring your ideas to life. Diverse style options within the github - bob-friedman protein-sequence-generation: python code to help collection suit various aesthetic preferences. Regular updates keep the github - bob-friedman protein-sequence-generation: python code to help collection current with contemporary trends and styles. Time-saving browsing features help users locate ideal github - bob-friedman protein-sequence-generation: python code to help images quickly. Comprehensive tagging systems facilitate quick discovery of relevant github - bob-friedman protein-sequence-generation: python code to help content. Instant download capabilities enable immediate access to chosen github - bob-friedman protein-sequence-generation: python code to help images.

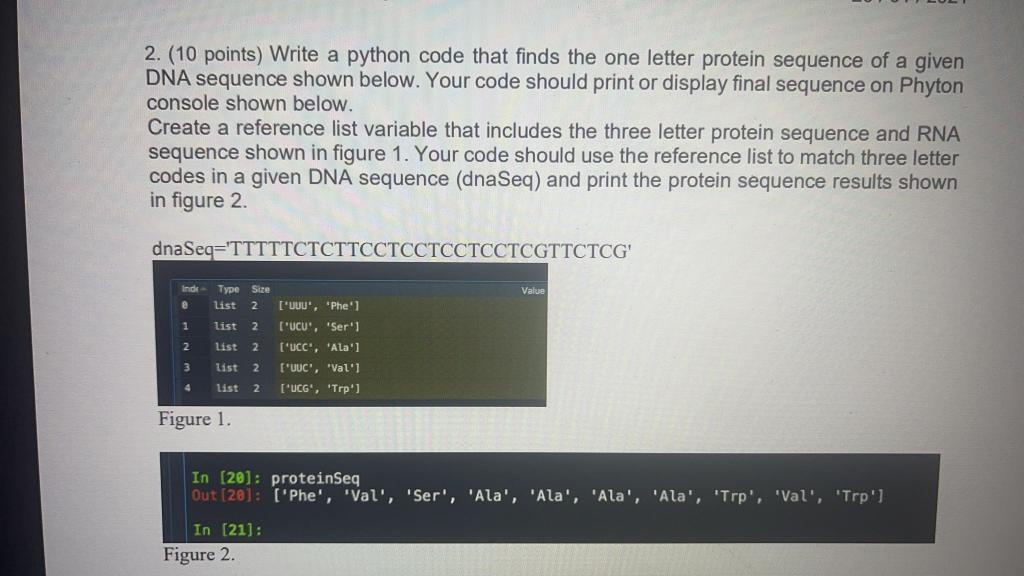








![Python code sequence [15] | Download Scientific Diagram](https://mavink.com/images/loadingwhitetransparent.gif)



















![[논문 리뷰] Secondary Structure-Guided Novel Protein Sequence Generation ...](https://moonlight-paper-snapshot.s3.ap-northeast-2.amazonaws.com/arxiv/secondary-structure-guided-novel-protein-sequence-generation-with-latent-graph-diffusion-1.png)




![[2403.03726] Diffusion on language model embeddings for protein ...](https://ar5iv.labs.arxiv.org/html/2403.03726/assets/figures/superfamily_fig_swissprot.png)


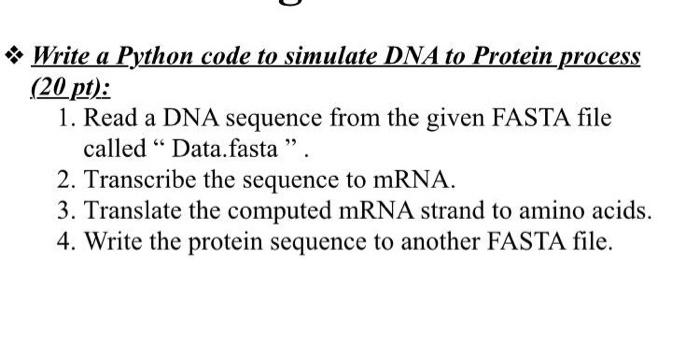
+corresponds+to+which+amino+acid.+2.7.S3+Use+a+table+of+mRNA+codons+and+their+corresponding+amino+acids+to+deduce+the+sequence+of+amino+acids+coded+by+a+short+mRNA+strand+of+known+base+sequence.+2.7.S4+Deducing+the+DNA+base+sequence+for+the+mRNA+strand..jpg)

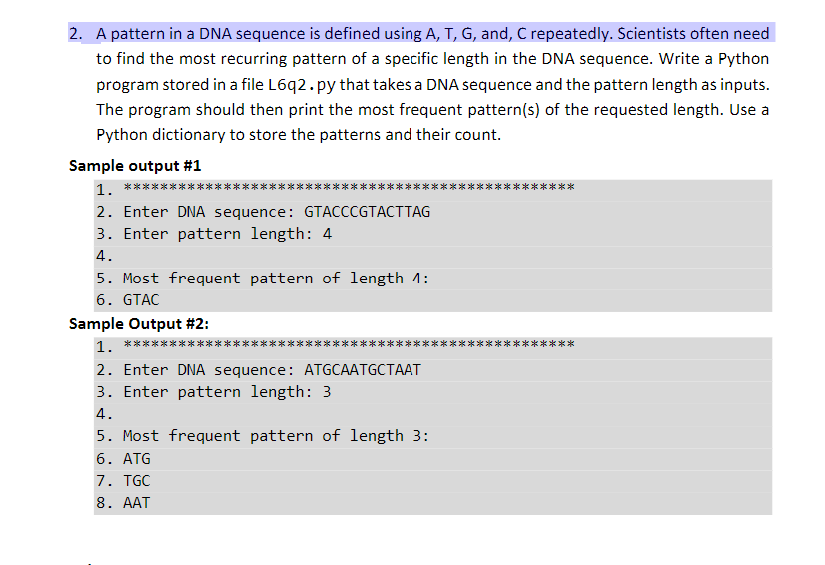




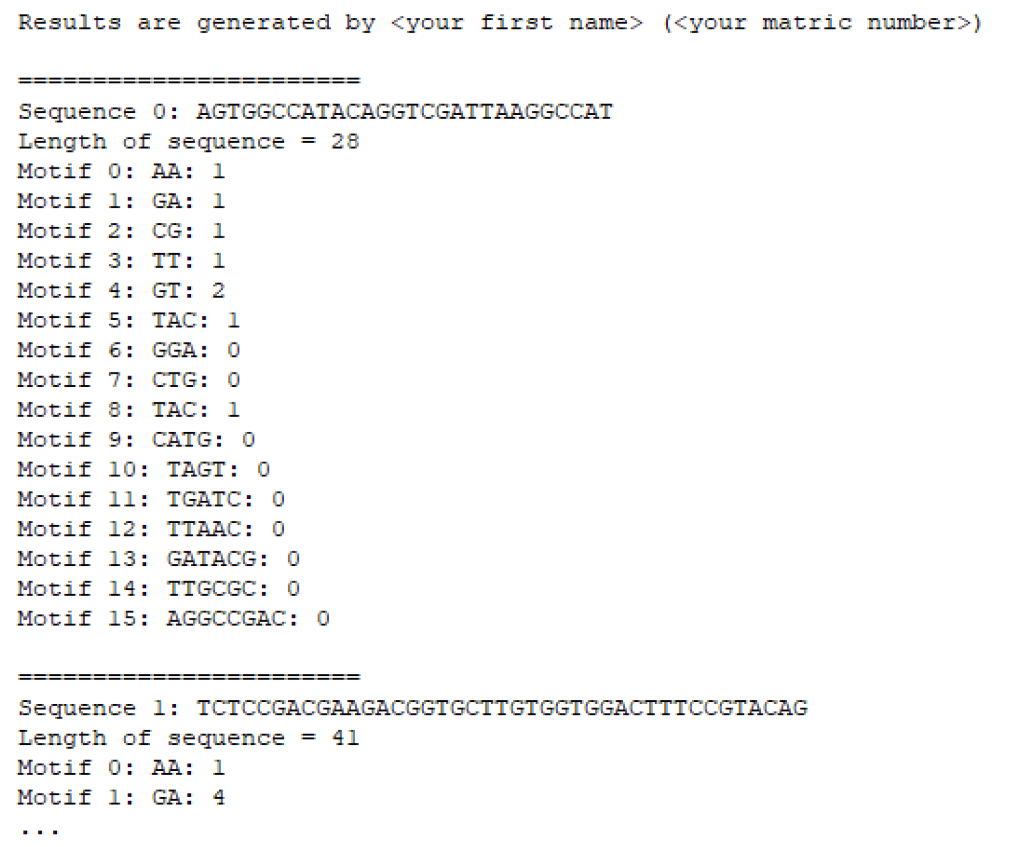
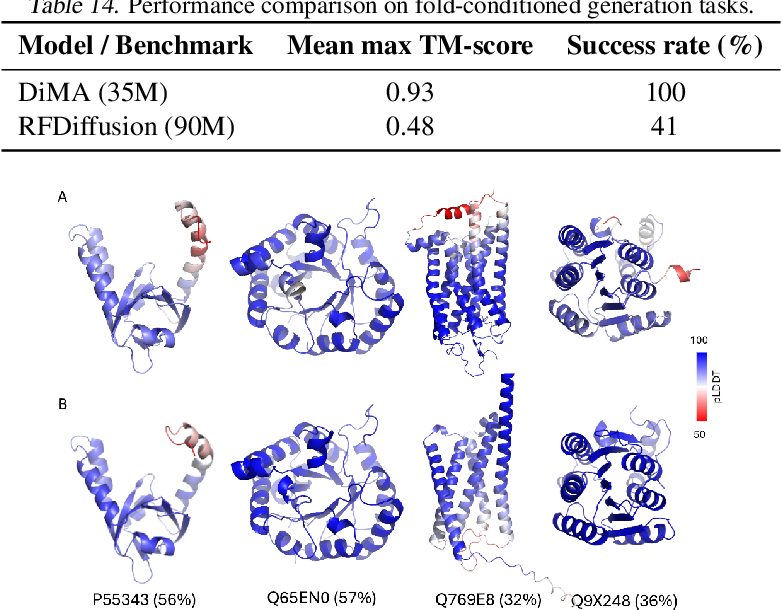
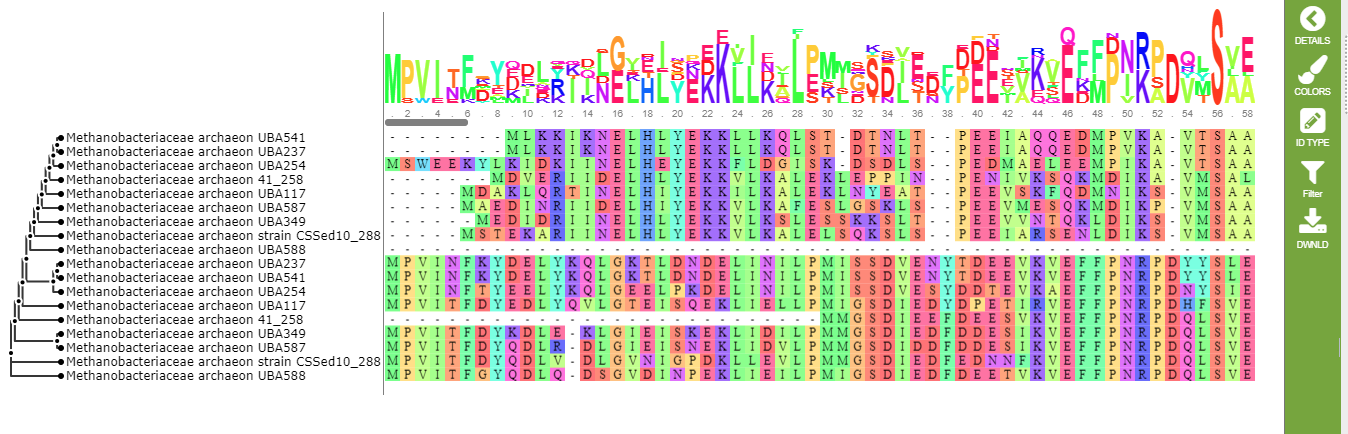













![PeptideBuilder: A simple Python library to generate model peptides [PeerJ]](https://peerj.com/articles/80/inlinefig-4.png)


